EMMAWiki/AdministratorDocumentation/ArraylayoutGuide: Difference between revisions
imported>MichaelDondrup No edit summary |
|||
| (11 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
= Arraylayout Guidelines = | = Arraylayout Guidelines = | ||
An arraylayout is essential for your microarray experiment. All further processing and analysis is based on the layout. A wrong layout leads to a wrong calculation! | == Synopsis == | ||
An Array layout describes the physical properties of a micorarray. '''An arraylayout is essential for your microarray experiment.''' All further processing and analysis is based on the layout. A wrong layout leads to a wrong calculation! Without a valid layout you will be unable to import any data. | |||
Making an Array layout available to EMMA is a two-step process: | |||
* Make an Array Description Format File (ADF) | |||
* Upload the ADF to EMMA providing few addtional data. | |||
Both steps can be performed by a Maintainer or Chief (that is possibly you :) ) and are described below. | |||
== Making an ADF == | |||
In order to provide MIAME compliant annotations, mandatory fields in the ADF format for EMMA are the same fields as in [http://www.ebi.ac.uk/arrayexpress/ ArrayExpress]. | In order to provide MIAME compliant annotations, mandatory fields in the ADF format for EMMA are the same fields as in [http://www.ebi.ac.uk/arrayexpress/ ArrayExpress]. | ||
| Line 13: | Line 25: | ||
You need to have the role ''Chief'' or ''Maintainer'' within your project to be able to upload ADF-files to EMMA. So check with the Maintainer of your project, to see how to upload your ADF. | You need to have the role ''Chief'' or ''Maintainer'' within your project to be able to upload ADF-files to EMMA. So check with the Maintainer of your project, to see how to upload your ADF. | ||
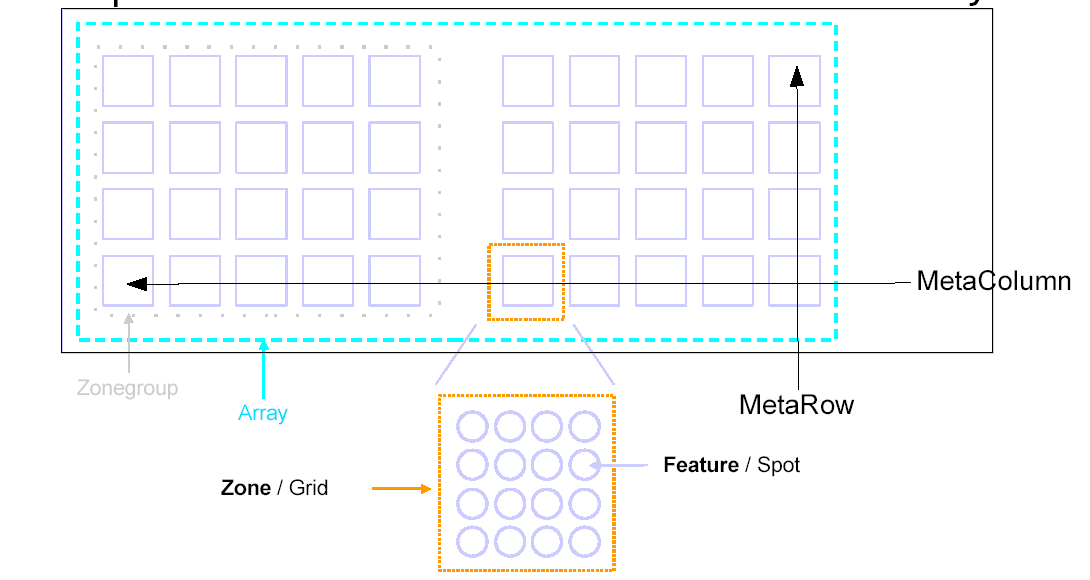
== Hints == | This graphic explains the most common terms used: | ||
[[File:arraydesign.png]] | |||
=== Hints === | |||
* The ADF file needs to be in '''text format'''. In Excel, use CSV export. | * The ADF file needs to be in '''text format'''. In Excel, use CSV export. | ||
| Line 19: | Line 35: | ||
* Quotes <code><nowiki>"" or ''</nowiki></code> around fields will not work with the ADF-checker | * Quotes <code><nowiki>"" or ''</nowiki></code> around fields will not work with the ADF-checker | ||
* Reporter identifiers must only consist of : '''A-Z,a-z,0-9,_,-,+,:''' In particular reporter identifiers may not contain: <code><nowiki> () tab [] ; </nowiki></code> | * Reporter identifiers must only consist of : '''A-Z,a-z,0-9,_,-,+,:''' In particular reporter identifiers may not contain: <code><nowiki> () tab [] ; </nowiki></code> | ||
* '''Do not try to conceil information''' on your array design (e.g. reporter sequences, annotation). You will need them later anyway. Let me tell you that the information you try to hide from the world is relevant only for yourself and your workgroup. It seems faulty to assume somebody in your project is going to steal your reporter sequence. | * '''Do not try to conceil information''' on your array design (e.g. reporter sequences, annotation). You will need them later anyway. Let me tell you that the information you try to hide from the world is relevant only for yourself and your workgroup. It seems faulty to assume somebody in your project is going to steal your reporter sequence. | ||
* '''Double check that Reporter Identifiers are correct.''' If you have the same reporter spotted in replicates, it needs to have the identical Reporter Identifier at that position. Reporter names or other details are irrelevant for this. | * '''Double check that Reporter Identifiers are correct.''' If you have the same reporter spotted in replicates, it needs to have the identical Reporter Identifier at that position. Reporter names or other details are irrelevant for this. | ||
* '''Use <code><nowiki>Reporter [[BioSequence]] Database Entry</nowiki></code> columns whenever possible.''' The entries will be shown as hyperlinks in the interface, if you provide a propper accession. You can provide multiple DB entry columns. Make sure the accession keys work when doing a manual query on the Database with your web-browser. Leave the field empty only if there is no db accession for this sequence. | * '''Use <code><nowiki>Reporter [[BioSequence]] Database Entry</nowiki></code> columns whenever possible.''' The entries will be shown as hyperlinks in the interface, if you provide a propper accession. You can provide multiple DB entry columns. Make sure the accession keys work when doing a manual query on the Database with your web-browser. Leave the field empty only if there is no db accession for this sequence. | ||
* If you have a list of BRIDGE links, that provide a external reference to a Sequence in GenDB or SAMS (aka.: <code><nowiki>o2xr://</nowiki></code>) you may use <code><nowiki>Reporter [[BioSequence]] Database Entry [GenDB]</nowiki></code> or <code><nowiki>Reporter [[BioSequence]] Database Entry [SAMS]</nowiki></code> to denote them. Instead of creating a regular Database entry, a BRIDGE reference for the Sequences is made. Include the full BRIDGE URI not only the id-number. | * If you have a list of BRIDGE links, that provide a external reference to a Sequence in GenDB or SAMS (aka.: <code><nowiki>o2xr://</nowiki></code>) you may use <code><nowiki>Reporter [[BioSequence]] Database Entry [GenDB]</nowiki></code> or <code><nowiki>Reporter [[BioSequence]] Database Entry [SAMS]</nowiki></code> to denote them. Instead of creating a regular Database entry, a BRIDGE reference for the Sequences is made. Include the full BRIDGE URI not only the id-number. | ||
| Line 27: | Line 43: | ||
* You can leave uncommon fields in you file as a reference and they will be appended to each Reporter description in the form <code><nowiki>[[FieldName]]=Value</nowiki></code>. | * You can leave uncommon fields in you file as a reference and they will be appended to each Reporter description in the form <code><nowiki>[[FieldName]]=Value</nowiki></code>. | ||
* Remember, an ADF file has to be uploaded only once for each [[ArrayLayout]] (or array type) not for each individual array. | * Remember, an ADF file has to be uploaded only once for each [[ArrayLayout]] (or array type) not for each individual array. | ||
== Uploading the ADF == | |||
# open the ''Advanced Actions'' menu | |||
# follow the link ''View arraylayouts'': | |||
[[File:leftmenu.png]] | |||
3.#3 click ''Import new arraylayout'' in Basic Actions: | |||
[[File:leftmenu1.png]] | |||
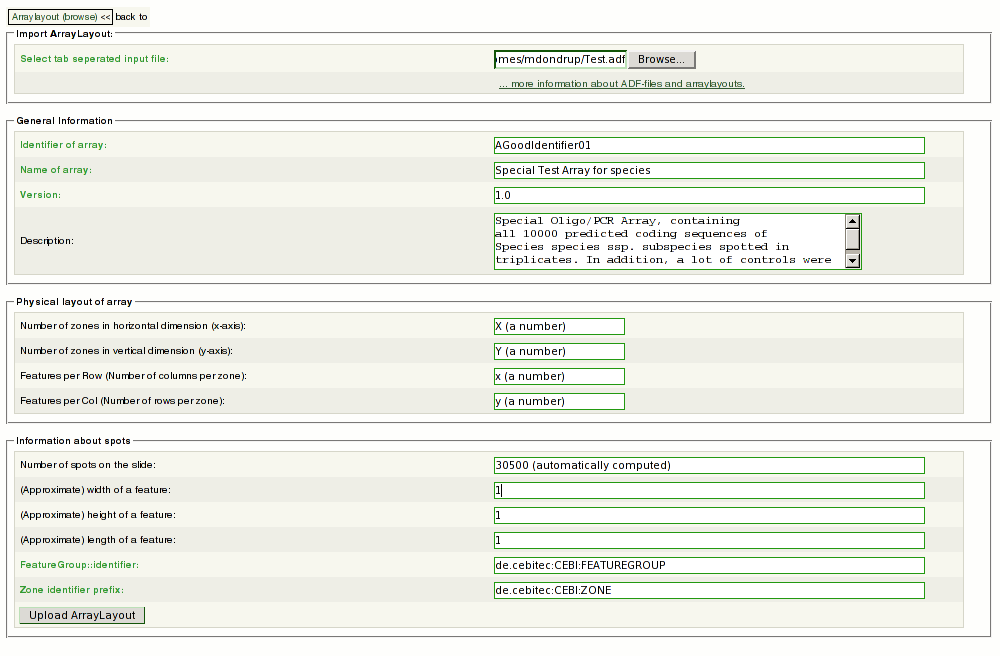
4.#4 Fill out the fields in the following form: | |||
[[File:arraylayoutUpload1.png]] | |||
<span style="font-size: x-large; color: red">!</span> Use the browse button to select the ADF on your local disc. You have to enter the mandatory information. | |||
Try to find a speaking name for your layout (''Mylayout'' for example is not a good idea). Even if not mandatory, you will need a good description. | |||
Remember: this is meant to aid only you and your colleagues. | |||
Enter the physical properties of your layout carefully : | |||
* a ''Zone = Grid'' in your layout | |||
* a ''Feature = Spot'' in your layout | |||
* ''horizontal axis'' is normally meant to refer the the shorter side (also called ''Metarows'', and ''Rows'') of the array | |||
* ''vertical axis'' refers to the longer side (''Metacolumns'' and ''Columns'') | |||
* EMMA will check if that conforms with the ADF you try to upload | |||
* The number of spots is calculated automatically from the above dimensions. | |||
Enter the other details if you know them. Leave the identifier prefix fields as they are. | |||
5.#5 When done, press the ''Upload [[ArrayLayout]]'' Button. EMMA will upload and check your layout. Please wait. | |||
This will take a few minutes to proceed. Often you will see an error message like this: [[File:arraylayoutUpload2.png]]. | |||
This means something is wrong with your ADF. The message is rather clear in what it does not like. Please correct the errors in your file and try again. | |||
If you messed only with the physical layout information, EMMA corrects the values automatically from parsing your file. If the values in red are correct you may still proceed. | |||
[[File:arraylayoutUpload3.png]] | |||
6.#6 You may review your settings. if everything is fine, click the ''Add to database'' button. | |||
This will create a background job that will add your layout permanently to your project. Follow the link to see the progress of your import. The import may take several hours. The [[ArrayLayout]] will be available only after it is finished. | |||
Latest revision as of 14:23, 28 October 2011
Arraylayout Guidelines
Synopsis
An Array layout describes the physical properties of a micorarray. An arraylayout is essential for your microarray experiment. All further processing and analysis is based on the layout. A wrong layout leads to a wrong calculation! Without a valid layout you will be unable to import any data.
Making an Array layout available to EMMA is a two-step process:
- Make an Array Description Format File (ADF)
- Upload the ADF to EMMA providing few addtional data.
Both steps can be performed by a Maintainer or Chief (that is possibly you :) ) and are described below.
Making an ADF
In order to provide MIAME compliant annotations, mandatory fields in the ADF format for EMMA are the same fields as in ArrayExpress.
Please have a look at the ADF Guidelines at the EBI for detailed descriptions and ADF Checklist at the EBI for a quick overview and checklist before you upload your ADF-file.
If you have a layout in the GAL-format, this can easily be converted to the ADF-format using the spotter file to ADF converter at the EBI. Do not expect to get a valid ADF file from this conversion. You will need to add additional mandatory columns.
Use the ADF checker provided by the EBI to interatively improve your file.
You need to have the role Chief or Maintainer within your project to be able to upload ADF-files to EMMA. So check with the Maintainer of your project, to see how to upload your ADF.
This graphic explains the most common terms used:
Hints
- The ADF file needs to be in text format. In Excel, use CSV export.
- Use only tab-delimited files
- Quotes
"" or ''around fields will not work with the ADF-checker - Reporter identifiers must only consist of : A-Z,a-z,0-9,_,-,+,: In particular reporter identifiers may not contain:
() tab [] ; - Do not try to conceil information on your array design (e.g. reporter sequences, annotation). You will need them later anyway. Let me tell you that the information you try to hide from the world is relevant only for yourself and your workgroup. It seems faulty to assume somebody in your project is going to steal your reporter sequence.
- Double check that Reporter Identifiers are correct. If you have the same reporter spotted in replicates, it needs to have the identical Reporter Identifier at that position. Reporter names or other details are irrelevant for this.
- Use
Reporter [[BioSequence]] Database Entrycolumns whenever possible. The entries will be shown as hyperlinks in the interface, if you provide a propper accession. You can provide multiple DB entry columns. Make sure the accession keys work when doing a manual query on the Database with your web-browser. Leave the field empty only if there is no db accession for this sequence. - If you have a list of BRIDGE links, that provide a external reference to a Sequence in GenDB or SAMS (aka.:
o2xr://) you may useReporter [[BioSequence]] Database Entry [GenDB]orReporter [[BioSequence]] Database Entry [SAMS]to denote them. Instead of creating a regular Database entry, a BRIDGE reference for the Sequences is made. Include the full BRIDGE URI not only the id-number. - Empty locations may not be ommited in the layout. Use the Reporter Identifier
Emptyto mark those locations. Set,Reporter Group [role]toControland ControlType tocontrol_empty. - Use the Reporter Comment field to add additional annotation to your Reporter. This will appear as a Reporter description in EMMA.
- You can leave uncommon fields in you file as a reference and they will be appended to each Reporter description in the form
[[FieldName]]=Value. - Remember, an ADF file has to be uploaded only once for each ArrayLayout (or array type) not for each individual array.
Uploading the ADF
- open the Advanced Actions menu
- follow the link View arraylayouts:
3.#3 click Import new arraylayout in Basic Actions:
4.#4 Fill out the fields in the following form:
! Use the browse button to select the ADF on your local disc. You have to enter the mandatory information. Try to find a speaking name for your layout (Mylayout for example is not a good idea). Even if not mandatory, you will need a good description. Remember: this is meant to aid only you and your colleagues.
Enter the physical properties of your layout carefully :
- a Zone = Grid in your layout
- a Feature = Spot in your layout
- horizontal axis is normally meant to refer the the shorter side (also called Metarows, and Rows) of the array
- vertical axis refers to the longer side (Metacolumns and Columns)
- EMMA will check if that conforms with the ADF you try to upload
- The number of spots is calculated automatically from the above dimensions.
Enter the other details if you know them. Leave the identifier prefix fields as they are.
5.#5 When done, press the Upload ArrayLayout Button. EMMA will upload and check your layout. Please wait.
This will take a few minutes to proceed. Often you will see an error message like this: 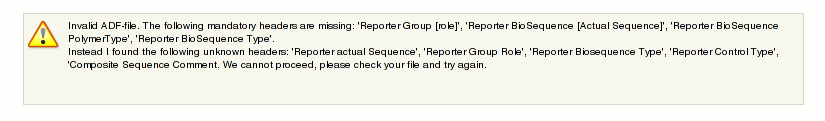 .
.
This means something is wrong with your ADF. The message is rather clear in what it does not like. Please correct the errors in your file and try again.
If you messed only with the physical layout information, EMMA corrects the values automatically from parsing your file. If the values in red are correct you may still proceed.
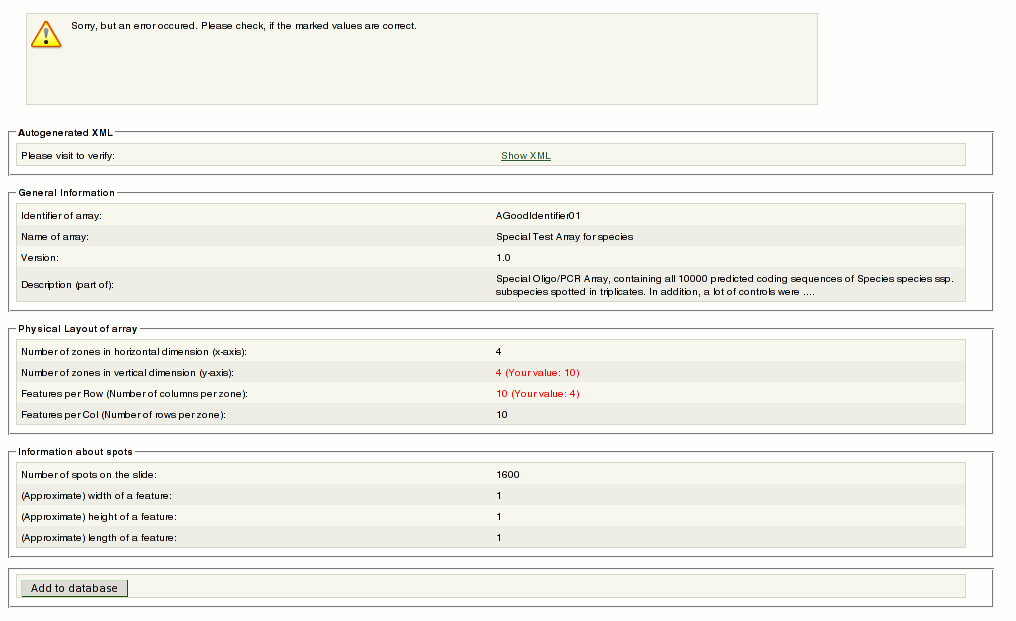
6.#6 You may review your settings. if everything is fine, click the Add to database button.
This will create a background job that will add your layout permanently to your project. Follow the link to see the progress of your import. The import may take several hours. The ArrayLayout will be available only after it is finished.