GenDBWiki/WebDocumentation/DialogWindows/SequenceViewer: Difference between revisions
imported>Keywan No edit summary |
No edit summary |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
= Sequence Viewer = | = Sequence Viewer = | ||
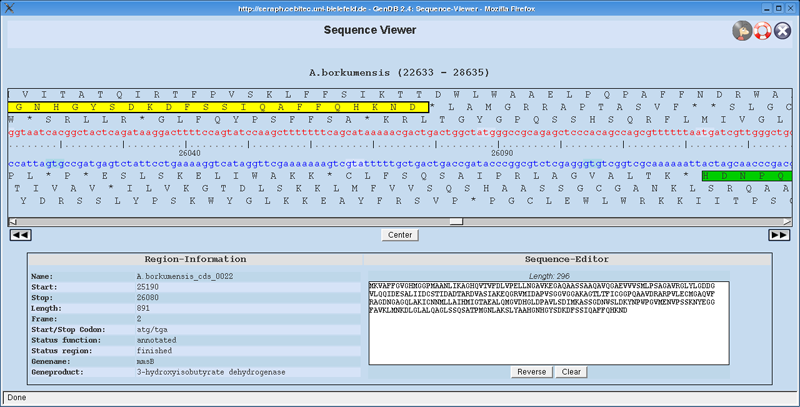
The Sequence Viewer (available since GenDB 2.4) allows a zoomed view into the genomic sequence along with its six translated frames. | |||
You can easily '''''select''''' some sequence, afterwards copy it with '''''Control+C''''', and finally paste it with '''''Control+V''''' into an arbitrary place. | |||
Pressing the Show Sequence button in the main window of GenDB, will display the [[SequenceViewer]]. Green rectangles represent the CDS. The CDS that is activated in GenDB has a stronger border and a brighter green color (directly reachable with the '''''Center''''' button). CDS that are marked as ignored are dashed and grey. Moving the mouse over a CDS, displays its details into the '''''Region-Information''''' table. The [[SequenceViewer]] allows you to scroll within a 6000nt-window. In order to get the next or previous 6000nt of the contig press the '''''Left''''' or '''''Right''''' button. Thus, its possible to walk easily through the whole contig. | |||
Selecting any sequence will display the selected sequence at the '''''Sequence-Editor'''''. If you select some sequence on the back strands, a '''''Reverse''''' button allows you to reverse the selected sequence, in order to obtain the sequence in the biological right direction. Afterwards you can select this sequence, copy and paste it into e.g. a file. | |||
=== How to select a sequence === | |||
The behaviour of the [[SequenceViewer]] is similar to common text-editors (e.g. notepad). You have two possibilities to select an arbitrary sequence: | |||
* Press the left mouse button and move it to the right or to the left (recommeneded for short sequences) | |||
* ''Left-Click'' on the start character, move the scroll bar, ''Shift+Left-Click'' on the end character (recommeneded for long sequences) | |||
In order to obtain quickly the whole sequence of a CDS: | |||
* Move the mouse to the left border of a CDS, ''Left-Click'' and pull the mouse slightly under | |||
[[File:SequenceViewer.png|Screenshot of the GenDB Sequence Viewer.]] | |||
Latest revision as of 11:19, 30 November 2011
Sequence Viewer
The Sequence Viewer (available since GenDB 2.4) allows a zoomed view into the genomic sequence along with its six translated frames. You can easily select some sequence, afterwards copy it with Control+C, and finally paste it with Control+V into an arbitrary place.
Pressing the Show Sequence button in the main window of GenDB, will display the SequenceViewer. Green rectangles represent the CDS. The CDS that is activated in GenDB has a stronger border and a brighter green color (directly reachable with the Center button). CDS that are marked as ignored are dashed and grey. Moving the mouse over a CDS, displays its details into the Region-Information table. The SequenceViewer allows you to scroll within a 6000nt-window. In order to get the next or previous 6000nt of the contig press the Left or Right button. Thus, its possible to walk easily through the whole contig.
Selecting any sequence will display the selected sequence at the Sequence-Editor. If you select some sequence on the back strands, a Reverse button allows you to reverse the selected sequence, in order to obtain the sequence in the biological right direction. Afterwards you can select this sequence, copy and paste it into e.g. a file.
How to select a sequence
The behaviour of the SequenceViewer is similar to common text-editors (e.g. notepad). You have two possibilities to select an arbitrary sequence:
- Press the left mouse button and move it to the right or to the left (recommeneded for short sequences)
- Left-Click on the start character, move the scroll bar, Shift+Left-Click on the end character (recommeneded for long sequences)
In order to obtain quickly the whole sequence of a CDS:
- Move the mouse to the left border of a CDS, Left-Click and pull the mouse slightly under