ProMeTraWiki/CebitecAccount: Difference between revisions
imported>HeikoNeuweger No edit summary |
No edit summary |
||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
= | = ProMeTra and CeBiTec accounts = | ||
Users of | Users of CeBiTec software resources such as [http://www.cebitec.uni-bielefeld.de/groups/brf/software/gendb GenDB], [http://www.cebitec.uni-bielefeld.de/groups/brf/software/gendb Emmma2], [http://qupe.cebitec.uni-bielefeld.de QuPE] or [http://meltdb.cebitec.uni-bielefeld.de MeltDB] can use their existing accounts to access the ProMeTra interface. This hast th advantage that only a single login procedure has to take place and afterwards experimental | ||
that only a single login procedure has to take place and afterwards experimental | data from the various ''Omics'' platforms of the [http://www.cebitec.uni-bielefeld.de/ CeBiTec] are made available via data exchange by WebService technology. | ||
data from the various '' | |||
[[File:CebitecAccount_login.png]] | |||
The | The main interface of ProMeTra currently allows to access Emma2, QuPE and MeltDB and users will | ||
see the experiments and datasets they already have access to in the individual applications. | |||
= Accessing datasets via the QuPE, Emma2 or MeltDB web services = | |||
[[File:CebitecAccount_webservices.png]] | |||
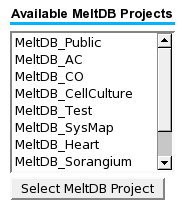
To request a dataset a user initially has to select a | |||
project. | |||
[[File:CebitecAccount_projects.png]] | |||
For the given project all available experiments are listed and ... | |||
[[File:CebitecAccount_experiments.png]] | |||
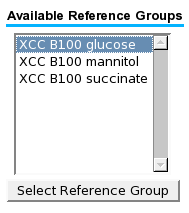
... a reference group or state can be selected afterwards. | |||
[[File:CebitecAccount_references.png]] | |||
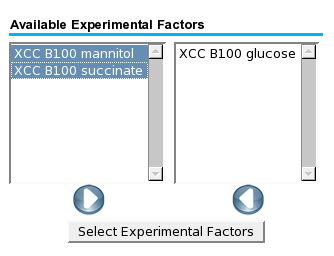
The groups or states that are selected in the last dialog will be compared to the | |||
references, and the resulting M-values will be used for the pathway or genome map visualizations. | |||
[[File:CebitecAccount_factors.png]] | |||
Latest revision as of 10:07, 21 March 2012
ProMeTra and CeBiTec accounts
Users of CeBiTec software resources such as GenDB, Emmma2, QuPE or MeltDB can use their existing accounts to access the ProMeTra interface. This hast th advantage that only a single login procedure has to take place and afterwards experimental data from the various Omics platforms of the CeBiTec are made available via data exchange by WebService technology.
The main interface of ProMeTra currently allows to access Emma2, QuPE and MeltDB and users will see the experiments and datasets they already have access to in the individual applications.
Accessing datasets via the QuPE, Emma2 or MeltDB web services
To request a dataset a user initially has to select a project.
For the given project all available experiments are listed and ...
... a reference group or state can be selected afterwards.
The groups or states that are selected in the last dialog will be compared to the references, and the resulting M-values will be used for the pathway or genome map visualizations.