ProMeTraWiki/GenomeMaps: Difference between revisions
imported>HeikoNeuweger No edit summary |
|||
| (6 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
= | = GenomeMaps in ProMeTra = | ||
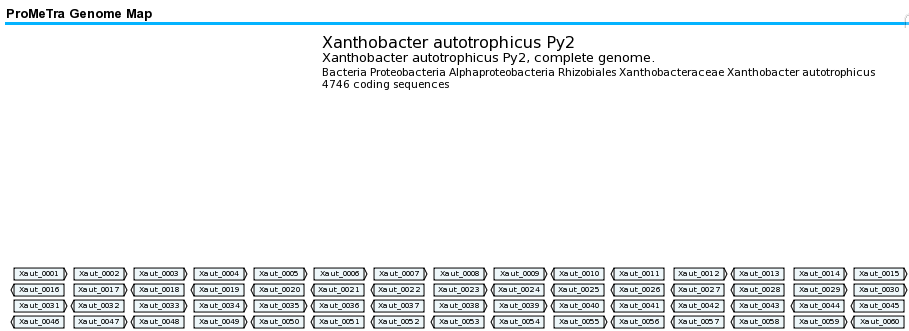
The replicons of prokaryotic genomes present in the NCBI genome database (as of Dez. 2008) | The replicons of prokaryotic genomes present in the NCBI genome database (as of Dez. 2008) have been converted into so called GenomeMaps for the ProMeTra web interface. A GenomeMap in the ProMeTra context is a SVG based representation of the coding sequences (CDS) identified and annotated in the GenBank files of the individual prokaryotic database entries. GenomeMaps represent CDS in a grid by individual rectangles labeled with the locus tag (or if available the gene name). The order of the CDSs in the Grid representation is determined by the basenumber of the stop codon counted from the origin of replication. The grid is filled by row, i.e. the first CDS is in first row, first column, the second CDS is located at first row second column and so forth. A triangle at the right side of a rectangle indicates that the corresponding CDS is located on the forward strand, a triangle to the left indicates the location on the reverse strand. | ||
have been converted into so called | |||
A | |||
individual prokaryotic database entries. | |||
The order of the CDSs in the Grid representation is determined by basenumber of the stop codon counted from the origin of replication. | |||
The grid is filled | |||
A triangle at the right side of a rectangle indicates that the corresponding CDS is located on the forward strand, a triangle to the left indicates reverse strand. | |||
Please see the following example of a GenomeMap for clarity: | |||
[[ | [[File:GenomeMaps_genomemap.png]] | ||
Latest revision as of 10:09, 21 March 2012
GenomeMaps in ProMeTra
The replicons of prokaryotic genomes present in the NCBI genome database (as of Dez. 2008) have been converted into so called GenomeMaps for the ProMeTra web interface. A GenomeMap in the ProMeTra context is a SVG based representation of the coding sequences (CDS) identified and annotated in the GenBank files of the individual prokaryotic database entries. GenomeMaps represent CDS in a grid by individual rectangles labeled with the locus tag (or if available the gene name). The order of the CDSs in the Grid representation is determined by the basenumber of the stop codon counted from the origin of replication. The grid is filled by row, i.e. the first CDS is in first row, first column, the second CDS is located at first row second column and so forth. A triangle at the right side of a rectangle indicates that the corresponding CDS is located on the forward strand, a triangle to the left indicates the location on the reverse strand.
Please see the following example of a GenomeMap for clarity: