MeltDBWiki/HowTos: Difference between revisions
Jump to navigation
Jump to search
imported>HeikoNeuweger No edit summary |
No edit summary |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
== Using the public MeltDB experiments == | == Using the public MeltDB experiments == | ||
Login to the public MeltDB project using the guest account: | * Login to the public MeltDB project using the guest account: | ||
[http://meltdb.cebitec.uni-bielefeld.de/cgi-bin/meltdb.cgi?login=guest&chksum=h/Zx4JEg6sz0I6hi97ZfsA&project=MeltDB_Public MeltDB guest login] | |||
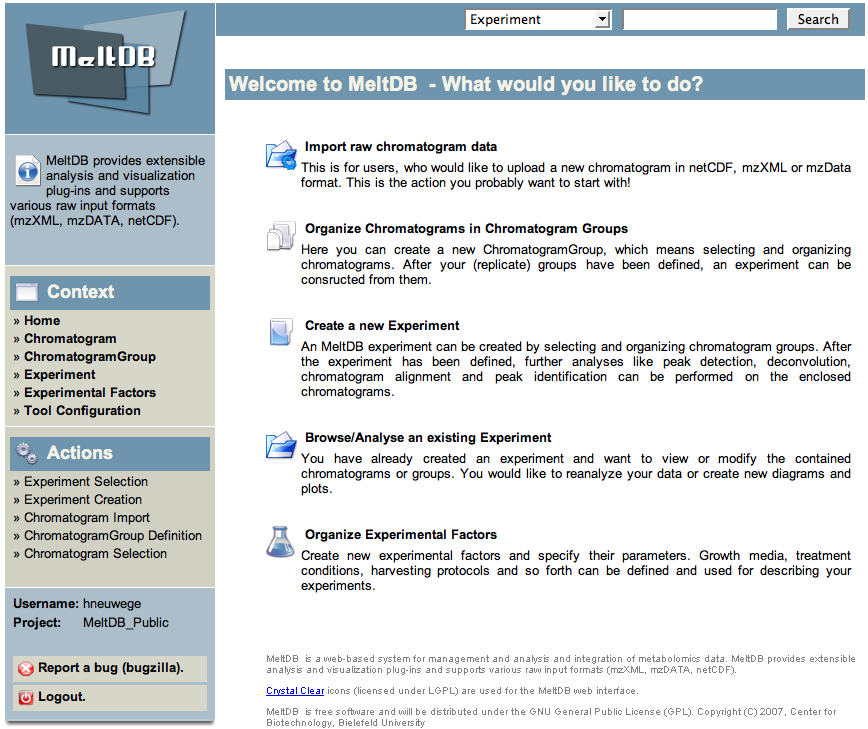
* To analyze the publicly available datasets in MeltDB, please select 'Browse/Analyse an existing Experiment' from the start page. | |||
[[Image:MeltDBWiki$$UploadChromas$startpage.png]] | |||
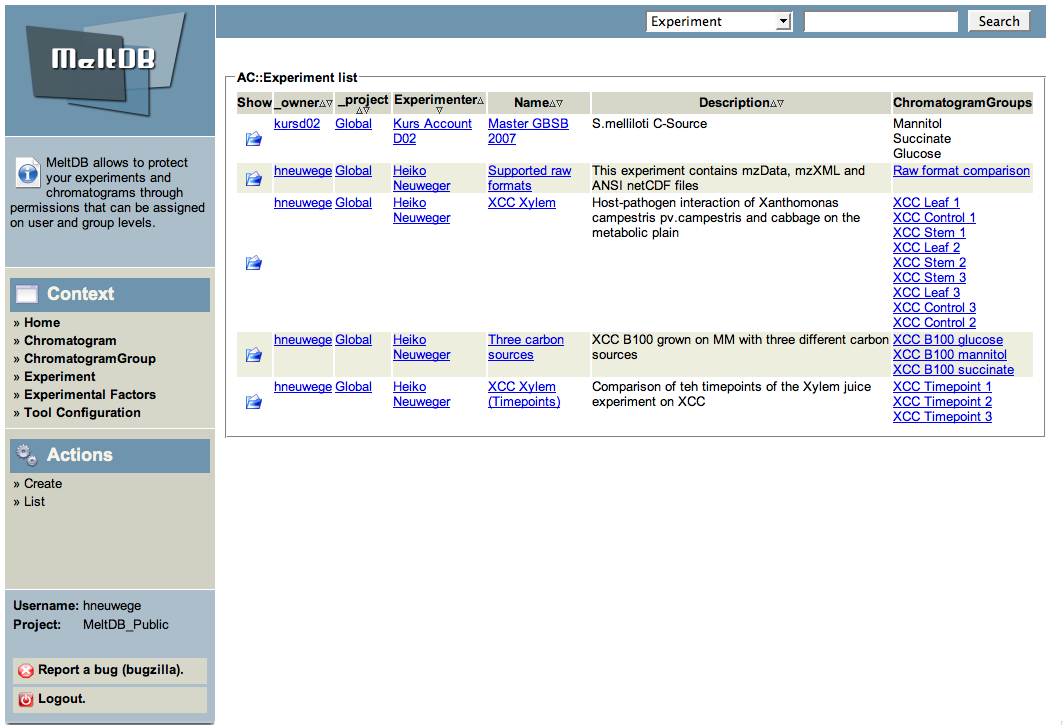
Select the 'Three carbon sources' experiment by clicking | * Select the 'Three carbon sources' experiment by clicking on the folder icon to the left. | ||
on the folder icon to the left. | |||
[[Image:MeltDBWiki$$HowTos$experiments.png]] | |||
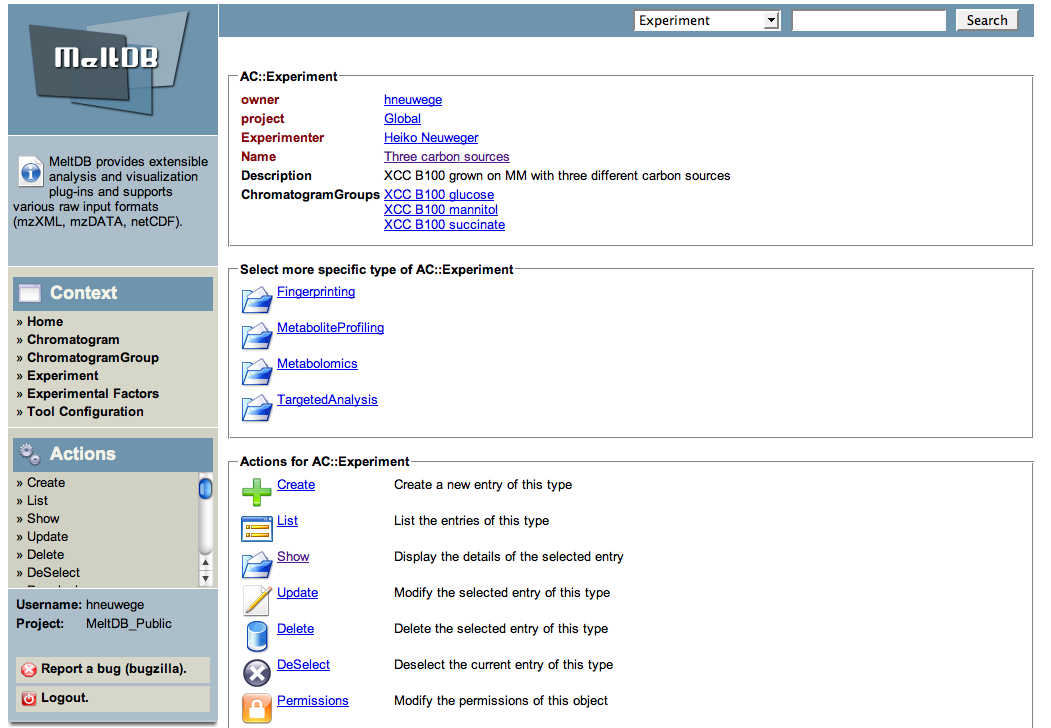
Select | * The textual display of the experiment is presented and a number of potential actions together with their short descriptions are listed below. | ||
[[Image:MeltDBWiki$$HowTos$carbon_sources.png]] | |||
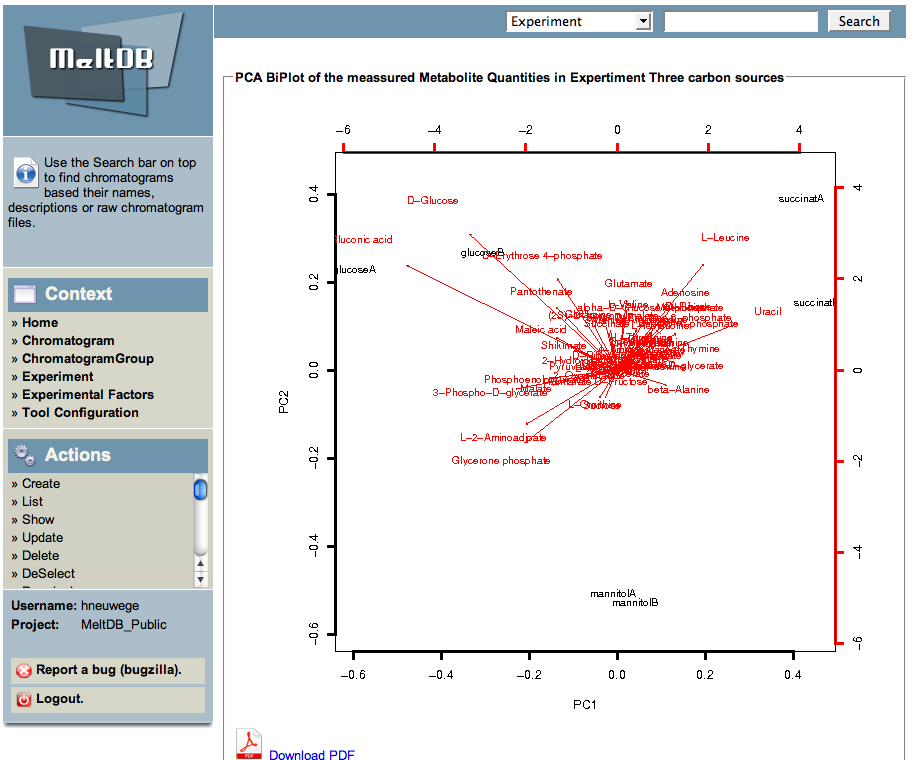
* Please select 'Show PCA' to start the PCA analysis on the previously imported peaks found by Xcalibur preprocessing methods. | |||
* Select all chromatograms and a list of metabolites and click the 'Compute PCA' button. If you would like to log-transform the measured and normalized metabolite values, mark the 'log' checkbox. | |||
[[Image:MeltDBWiki$$HowTos$pca.png]] | |||
Latest revision as of 16:12, 31 October 2011
Using the public MeltDB experiments
- Login to the public MeltDB project using the guest account:
MeltDB guest login
- To analyze the publicly available datasets in MeltDB, please select 'Browse/Analyse an existing Experiment' from the start page.
- Select the 'Three carbon sources' experiment by clicking on the folder icon to the left.
- The textual display of the experiment is presented and a number of potential actions together with their short descriptions are listed below.
- Please select 'Show PCA' to start the PCA analysis on the previously imported peaks found by Xcalibur preprocessing methods.
- Select all chromatograms and a list of metabolites and click the 'Compute PCA' button. If you would like to log-transform the measured and normalized metabolite values, mark the 'log' checkbox.