MeltDBWiki/PerformFBCA: Difference between revisions
Jump to navigation
Jump to search
imported>HeikoNeuweger No edit summary |
imported>HeikoNeuweger No edit summary |
||
| Line 8: | Line 8: | ||
a) | a) | ||
[[Image:MeltDBWiki$$PerformFBCA$browse.png]] | |||
b) | b) | ||
[[Image:MeltDBWiki$$PerformFBCA$search.png]] | |||
* After selecting your experiment, you can generate a multiple TIC view of the associated chromatograms via the following function: | * After selecting your experiment, you can generate a multiple TIC view of the associated chromatograms via the following function: | ||
[[Image:MeltDBWiki$$PerformFBCA$tic2.png]] | |||
* The first call to this may take some time since the image needs to be rendered from the raw datasets. Consecutive calls will reuse the generated image. If you change your experiment by adding or removing chromatograms to the chromatogram groups you can repaint the image with the 'recreate image' link. | * The first call to this may take some time since the image needs to be rendered from the raw datasets. Consecutive calls will reuse the generated image. If you change your experiment by adding or removing chromatograms to the chromatogram groups you can repaint the image with the 'recreate image' link. | ||
[[Image:MeltDBWiki$$PerformFBCA$tic.png]] | |||
* Feature based alignment is only possible if a peak detection and/or import was performed on your chromatograms. In order to compute an alignment using the FBCA tool of MeltDB select a representative chromatogram of your experiment. (e.g. click on one peak marked as blue or green spot and select the chromatogram name below). Select the 'Compute aligment' action from the chromatogram context menu. | * Feature based alignment is only possible if a peak detection and/or import was performed on your chromatograms. In order to compute an alignment using the FBCA tool of MeltDB select a representative chromatogram of your experiment. (e.g. click on one peak marked as blue or green spot and select the chromatogram name below). Select the 'Compute aligment' action from the chromatogram context menu. | ||
[[Image:MeltDBWiki$$PerformFBCA$fbca1.png]] | |||
* The FBCA algorithm will generate pairwise alignments of your selected chromatogram with all other chromatograms of the selected experiment. Click 'Compute aligment' to start the processing. This may take some time... | * The FBCA algorithm will generate pairwise alignments of your selected chromatogram with all other chromatograms of the selected experiment. Click 'Compute aligment' to start the processing. This may take some time... | ||
[[Image:MeltDBWiki$$PerformFBCA$fbca2.png]] | |||
* After the computation is finished, use the 'Show alignment' action of your chromatogram to review the results. You can reorder the chromatograms and/or select only subsets of the aligned chromatograms in the dialog. Select the peak detection or importer tools for which you would like to view peaks in the alignment. | * After the computation is finished, use the 'Show alignment' action of your chromatogram to review the results. You can reorder the chromatograms and/or select only subsets of the aligned chromatograms in the dialog. Select the peak detection or importer tools for which you would like to view peaks in the alignment. | ||
[[Image:MeltDBWiki$$PerformFBCA$fbca3.png]] | |||
* The rendering of the aligned image takes some seconds, it allows to quickly spot differences in the aligned chromatograms. | * The rendering of the aligned image takes some seconds, it allows to quickly spot differences in the aligned chromatograms. | ||
[[Image:MeltDBWiki$$PerformFBCA$fbca4.png]] | |||
Revision as of 15:42, 23 September 2008
Perform the Feature Based Chromatogram Alignment of MeltDB
- After logging in to MeltDB, select your experiment by either a) browsing through the list of available experiments or b) via the search function available at the top.
a)
b)
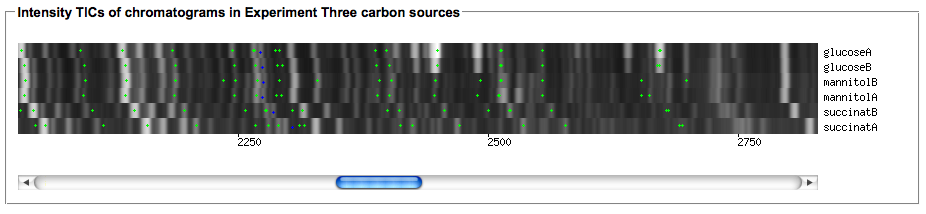
- After selecting your experiment, you can generate a multiple TIC view of the associated chromatograms via the following function:
- The first call to this may take some time since the image needs to be rendered from the raw datasets. Consecutive calls will reuse the generated image. If you change your experiment by adding or removing chromatograms to the chromatogram groups you can repaint the image with the 'recreate image' link.
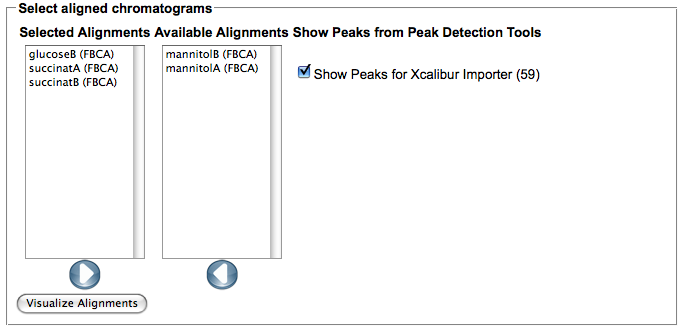
- Feature based alignment is only possible if a peak detection and/or import was performed on your chromatograms. In order to compute an alignment using the FBCA tool of MeltDB select a representative chromatogram of your experiment. (e.g. click on one peak marked as blue or green spot and select the chromatogram name below). Select the 'Compute aligment' action from the chromatogram context menu.
- The FBCA algorithm will generate pairwise alignments of your selected chromatogram with all other chromatograms of the selected experiment. Click 'Compute aligment' to start the processing. This may take some time...
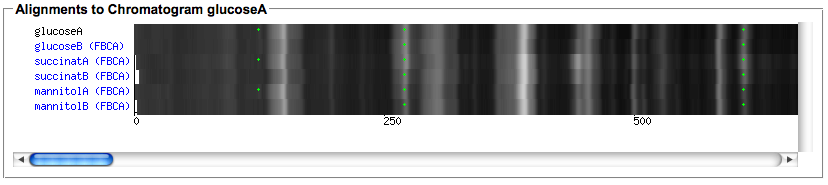
- After the computation is finished, use the 'Show alignment' action of your chromatogram to review the results. You can reorder the chromatograms and/or select only subsets of the aligned chromatograms in the dialog. Select the peak detection or importer tools for which you would like to view peaks in the alignment.
- The rendering of the aligned image takes some seconds, it allows to quickly spot differences in the aligned chromatograms.