GenDBWiki/DeveloperDocumentation/SystemDesign: Difference between revisions
(Created page with "= The GenDB System Design = The figure below illustrates the architecture of the GenDB system with the main building blocks. attachment:GenDB-Overview.png On the server side ...") |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 4: | Line 4: | ||
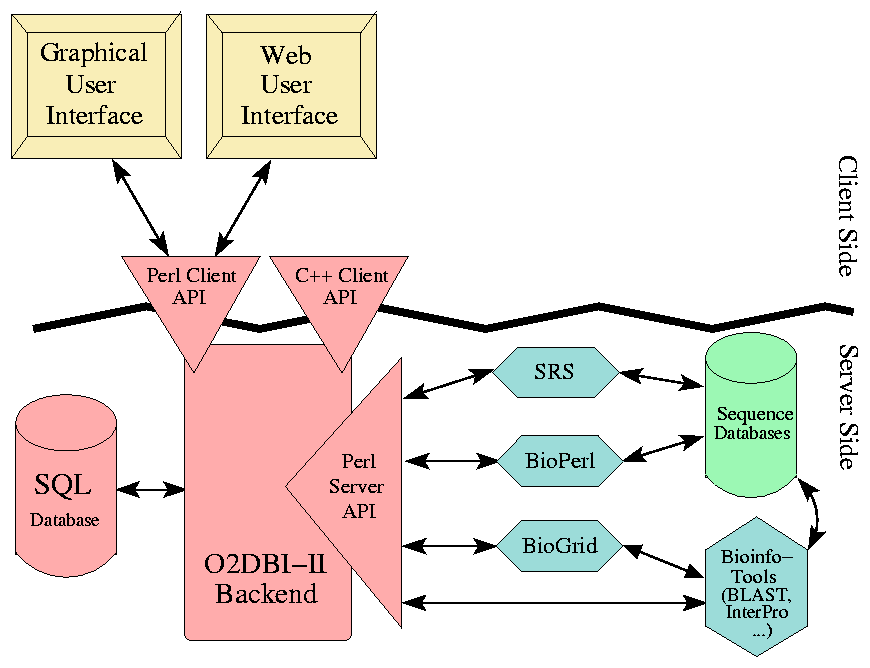
The figure below illustrates the architecture of the GenDB system with the main building blocks. | The figure below illustrates the architecture of the GenDB system with the main building blocks. | ||
[[File:GenDB-Overview.png]] | |||
On the server side all data that is stored in a relational database management system can be accessed using the O2DBI modules. The Perl server API is basically used for the integration of sequence databases and bioinformatics tools. On the client side, different user frontends are implemented that access the system via a Perl or C++ client API. | On the server side all data that is stored in a relational database management system can be accessed using the O2DBI modules. The Perl server API is basically used for the integration of sequence databases and bioinformatics tools. On the client side, different user frontends are implemented that access the system via a Perl or C++ client API. | ||
| Line 10: | Line 10: | ||
The complexity of the system encourages using an object-oriented approach not only in designing but also in implementing the system. Therefore, the enhanced O2DBI system was used to map Perl objects automatically to relational tables. The GenDB objects are mapped onto tables via O2DBI and stored in a relational database (here [http://www.mysql.com/ MySQL]). All access to the data via a Perl client or server API, or via a C++ client interface is managed again by the O2DBI module. | The complexity of the system encourages using an object-oriented approach not only in designing but also in implementing the system. Therefore, the enhanced O2DBI system was used to map Perl objects automatically to relational tables. The GenDB objects are mapped onto tables via O2DBI and stored in a relational database (here [http://www.mysql.com/ MySQL]). All access to the data via a Perl client or server API, or via a C++ client interface is managed again by the O2DBI module. | ||
On the client side, user interfaces have been implemented that use the functionality of these APIs. On the server side, sequence databases can be accessed via the SRS system or via the | On the client side, user interfaces have been implemented that use the functionality of these APIs. On the server side, sequence databases can be accessed via the SRS system or via the BioPerl interfaces. Computation-intensive tools like Blast or !InterPro can be managed and scheduled via a BioGrid (e.g. [http://www.oracle.com/ Oracle GridEngine]). | ||
Latest revision as of 18:23, 31 October 2011
The GenDB System Design
The figure below illustrates the architecture of the GenDB system with the main building blocks.
On the server side all data that is stored in a relational database management system can be accessed using the O2DBI modules. The Perl server API is basically used for the integration of sequence databases and bioinformatics tools. On the client side, different user frontends are implemented that access the system via a Perl or C++ client API.
The complexity of the system encourages using an object-oriented approach not only in designing but also in implementing the system. Therefore, the enhanced O2DBI system was used to map Perl objects automatically to relational tables. The GenDB objects are mapped onto tables via O2DBI and stored in a relational database (here MySQL). All access to the data via a Perl client or server API, or via a C++ client interface is managed again by the O2DBI module.
On the client side, user interfaces have been implemented that use the functionality of these APIs. On the server side, sequence databases can be accessed via the SRS system or via the BioPerl interfaces. Computation-intensive tools like Blast or !InterPro can be managed and scheduled via a BioGrid (e.g. Oracle GridEngine).