GenDBWiki/TermsAndConcepts/GenDBAnnotationWorkflow
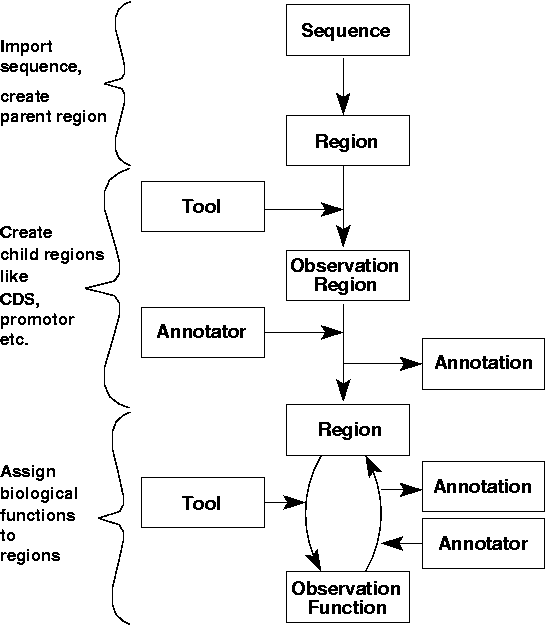
A typical GenDB Annotation Workflow
The GenDB-2 system features all steps for the analysis and annotation of bacterial genomes starting from the raw contig sequence. The figure below shows an example for a genome annotation pipeline that has been implemented with GenDB. Upon import of the raw sequence data, a parent region object describing the genome sequence is created. Following this step, user-defined tools for the prediction of different kinds of regions, such as coding sequences (CDS) or tRNA-encoding genes can be run. The output of these tools is stored as observations which refer to the parent region object. Based on these observations, an annotator, human or machine, performs a region annotation. This means confirming or rejecting the results of gene prediction tools by creating region objects like CDSs or tRNAs. The annotations form a complete protocol of all region annotation events. Following the creation of different kinds of regions, additional tools such as BLAST, HMMer, or !CoBias can be run creating information related to their potential function. Each of these tools can have its own automatic annotator that creates a very simple annotation based solely on the results of a single tool run. After computing a number of standard tools, a more sophisticated automatic annotation can be accomplished by combining the results of different tools. Finally, a manual function annotation step can be performed by an annotator in which a putative function is assigned to these regions by an interpretation of the observations (see below).
This standard sample pipeline implemented with GenDB-2 starts with an import of a contig sequence. Afterwards, regions are predicted and created by a regional annotation (Annotation::Region). A biological function for these regions can then be assigned by computing different bioinformatics tools that often generate large numbers of observations. Based on these results an automatic or manual functional annotation (Annotation::Function) can be assigned.
The current Gtk version of the GenDB system features a graphical interface (Annotation Pipeline Wizard) for the configuration of different individual pipelines. The user can choose one or more steps (Import, Edit Sequence, Region Prediction and Function Prediction) which are then combined to a separate pipeline. After some initial configuration, the pipeline is submitted as a special job and the corresponding steps are executed in the specified order without any further user interaction. Using these pipelines allows a very comfortable automated annotation and increases the productivity in large-scale genome annotation projects.
Nevertheless, it is still a laborious task to manually check the predicted regions and their function assignments. Both GenDB frontends therefore provide almost identical wizards for editing the start of a gene and annotation interfaces that allow recording a comprehensive set of information about each region. Since the final manual annotation of a genome is not only the most time consuming step but also the most erroneous task, exactly defined rules and guidelines for annotating a gene are essential in order to prevent inconsistent entries.