GenDBWiki/WebDocumentation/DialogWindows/AnnotationDialog: Difference between revisions
(Created page with "= Annotation Dialog = In this section the manual annotation of a Region using the Web Interface is described. A description of the terms and concepts of the GenDB annotation co...") |
No edit summary |
||
| Line 2: | Line 2: | ||
In this section the manual annotation of a Region using the Web Interface is | In this section the manual annotation of a Region using the Web Interface is | ||
described. A description of the terms and concepts of the GenDB annotation | described. A description of the terms and concepts of the GenDB annotation | ||
concept can be found in the [ | concept can be found in the [[GenDBWiki/TermsAndConcepts/AnnotationConcepts|Annotation Concepts]] of the [[GenDBWiki/TermsAndConcepts|Terms and Concepts Section]] | ||
Two kinds of annotations do fully characterize a region: | Two kinds of annotations do fully characterize a region: | ||
* A Region Annotation | |||
* A Function Annotation | |||
== Region Annotation == | == Region Annotation == | ||
The following image shows the annotation dialog of the GenDB Web interface. | The following image shows the annotation dialog of the GenDB Web interface. | ||
The first line informs about the region name and - if available - the biological | The first line informs about the region name and - if available - the biological | ||
name of the region. On both sides of this line there are arrows to access the | name of the region. On both sides of this line there are arrows to access the | ||
previous or the following of the current region. | previous or the following of the current region. | ||
On the left side, the list of functional and regional annotations is located. | On the left side, the list of functional and regional annotations is located. | ||
The selected annotation is marked in blue and shown on the right side in the | The selected annotation is marked in blue and shown on the right side in the | ||
Annotation Detail section. In this case, it is a regional annotation done by the | Annotation Detail section. In this case, it is a regional annotation done by the | ||
Critica auto annotator. | Critica auto annotator. | ||
| Line 30: | Line 29: | ||
Following the date and the annotator (which can be a human or an autoannotator) the | Following the date and the annotator (which can be a human or an autoannotator) the | ||
start and the stop position of the region are displayed. These can be changed in the | start and the stop position of the region are displayed. These can be changed in the | ||
[ | [[GenDBWiki/WebDocumentation/DialogWindows/RegionEditor|Region Editor]]. | ||
After the description field and the comment field there are the observations that | After the description field and the comment field there are the observations that | ||
| Line 41: | Line 40: | ||
On the bottom and on the top of the Annotation Dialog there are three buttons, respectively. | On the bottom and on the top of the Annotation Dialog there are three buttons, respectively. | ||
These have the functions to add a new annotation ('Add new') or accept an annotation (which | These have the functions to add a new annotation ('Add new') or accept an annotation (which | ||
means the same as 'Add new' but the regional status is automatically set to 'finished' and | means the same as 'Add new' but the regional status is automatically set to 'finished' and | ||
the functional status is automatically set to 'annotated'). To make no changes and just | the functional status is automatically set to 'annotated'). To make no changes and just | ||
close the annotation dialog press the 'Close' button. | close the annotation dialog press the 'Close' button. | ||
attachment:annotationDialogRegion.png | attachment:annotationDialogRegion.png | ||
== Function Annotation == | == Function Annotation == | ||
Choosing a functional annotation on the left side a number of additional fields occur on | Choosing a functional annotation on the left side a number of additional fields occur on | ||
the right side. One additional field is an entry for the | the right side. One additional field is an entry for the gene name for coding regions which | ||
is a biological identifier for the region. A certain gene name should only be given once | is a biological identifier for the region. A certain gene name should only be given once | ||
for a genome. | for a genome. | ||
Right from the | Right from the gene name entry there is the possibility to add GeneOntology numbers as a | ||
functional classification for the gene. More information to each GO number can be accessed | functional classification for the gene. More information to each GO number can be accessed | ||
via the links next to the field. | via the links next to the field. | ||
The EC-Number entry can contain one Enzyme Commission number that is a numerical classification | The EC-Number entry can contain one Enzyme Commission number that is a numerical classification | ||
scheme for enzymes. Genes that do not code for enzymes do not get such a number. The EC numbers | scheme for enzymes. Genes that do not code for enzymes do not get such a number. The EC numbers | ||
can be used for the reconstruction of the metabolic pathways of the organism e.g. in | can be used for the reconstruction of the metabolic pathways of the organism e.g. in [[GenDBWiki/WebDocumentation/MainViews/KEGGView|KEGG]]. | ||
[ | |||
In the Gene Product field the gene is shortly characterized. If there is no exact characterization | In the Gene Product field the gene is shortly characterized. If there is no exact characterization | ||
possible, there are different types of classes available for a gene product. These can be | possible, there are different types of classes available for a gene product. These can be | ||
accessed in the drop down menu below the Gene Product entry and is entered in the entry after | accessed in the drop down menu below the Gene Product entry and is entered in the entry after | ||
choosing one. Examples are 'hypothetical protein' if no further information is available for | choosing one. Examples are 'hypothetical protein' if no further information is available for | ||
the gene product or 'conserved hypothetical protein' if there is at least one good hit to | the gene product or 'conserved hypothetical protein' if there is at least one good hit to | ||
a protein in another organism. | a protein in another organism. | ||
| Line 77: | Line 75: | ||
Below the description field there is the check box 'Experimental' which should be clicked if any | Below the description field there is the check box 'Experimental' which should be clicked if any | ||
of the results of the annotation are verified in the wet lab. Following are entries concerning | of the results of the annotation are verified in the wet lab. Following are entries concerning | ||
the COG (Cluster of orthologous group) functional classification, namely the COG number, COG | the COG (Cluster of orthologous group) functional classification, namely the COG number, COG | ||
funccat ID and COG funccat Description. These fields cannot be edited manually, but only | funccat ID and COG funccat Description. These fields cannot be edited manually, but only | ||
pressing the button 'Get' on the right. In the upcoming dialog a COG number can be selected and | pressing the button 'Get' on the right. In the upcoming dialog a COG number can be selected and | ||
the entries will be filled. | the entries will be filled. | ||
In the 'Function' field a short description of the function of the gene can be inserted. | In the 'Function' field a short description of the function of the gene can be inserted. | ||
A confidence for the complete annotation is also to be entered in the respective drop down field. | A confidence for the complete annotation is also to be entered in the respective drop down field. | ||
There are different confidence stati that can be chosen here. The strongest confidence is | There are different confidence stati that can be chosen here. The strongest confidence is | ||
'High confidence in function and specificity' and the weakest is 'Hypothetical protein'. | 'High confidence in function and specificity' and the weakest is 'Hypothetical protein'. | ||
The following fields have already been described for the | The following fields have already been described for the Region Annotation. | ||
[[File:annotationDialog.png]] | [[File:annotationDialog.png]] | ||
Revision as of 21:55, 15 November 2011
Annotation Dialog
In this section the manual annotation of a Region using the Web Interface is described. A description of the terms and concepts of the GenDB annotation concept can be found in the Annotation Concepts of the Terms and Concepts Section
Two kinds of annotations do fully characterize a region:
- A Region Annotation
- A Function Annotation
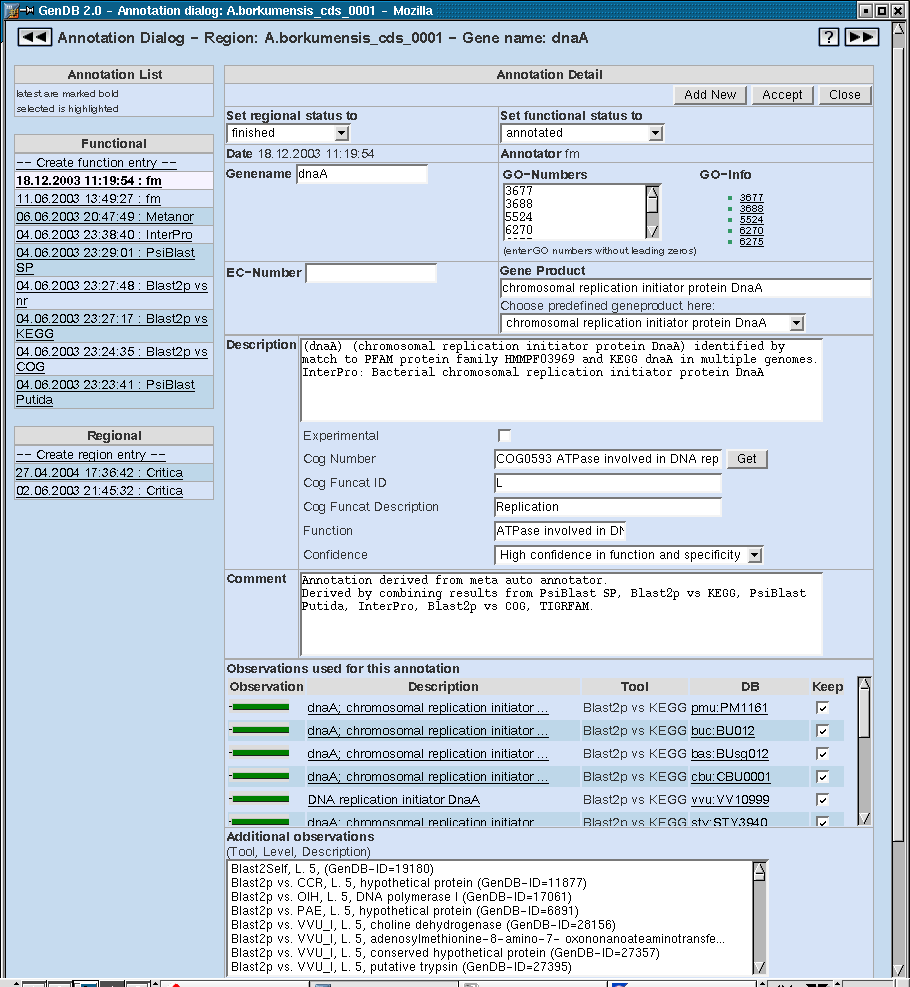
Region Annotation
The following image shows the annotation dialog of the GenDB Web interface. The first line informs about the region name and - if available - the biological name of the region. On both sides of this line there are arrows to access the previous or the following of the current region.
On the left side, the list of functional and regional annotations is located. The selected annotation is marked in blue and shown on the right side in the Annotation Detail section. In this case, it is a regional annotation done by the Critica auto annotator.
In the two drop down menues 'Set regional status to' and 'Set functional status to' a number of stati for the two different annotation types can be chosen. A completely annotated coding sequence should have the two stati shown here which is 'finished' for the regional and 'annotated' for the functional annotation.
Following the date and the annotator (which can be a human or an autoannotator) the start and the stop position of the region are displayed. These can be changed in the Region Editor.
After the description field and the comment field there are the observations that leaded to the current annotation listed. The following field contains additional observations that can be added to the list via the buttons 'Show selected ...' to look at selected observations or 'Add observations...' to add them manually to the list.
The last section shows links for the current annotation. New links can be added in the field right to the links.
On the bottom and on the top of the Annotation Dialog there are three buttons, respectively. These have the functions to add a new annotation ('Add new') or accept an annotation (which means the same as 'Add new' but the regional status is automatically set to 'finished' and the functional status is automatically set to 'annotated'). To make no changes and just close the annotation dialog press the 'Close' button.
attachment:annotationDialogRegion.png
Function Annotation
Choosing a functional annotation on the left side a number of additional fields occur on the right side. One additional field is an entry for the gene name for coding regions which is a biological identifier for the region. A certain gene name should only be given once for a genome.
Right from the gene name entry there is the possibility to add GeneOntology numbers as a functional classification for the gene. More information to each GO number can be accessed via the links next to the field.
The EC-Number entry can contain one Enzyme Commission number that is a numerical classification scheme for enzymes. Genes that do not code for enzymes do not get such a number. The EC numbers can be used for the reconstruction of the metabolic pathways of the organism e.g. in KEGG.
In the Gene Product field the gene is shortly characterized. If there is no exact characterization possible, there are different types of classes available for a gene product. These can be accessed in the drop down menu below the Gene Product entry and is entered in the entry after choosing one. Examples are 'hypothetical protein' if no further information is available for the gene product or 'conserved hypothetical protein' if there is at least one good hit to a protein in another organism.
The Description field of the functional annotation is a further detailed characterization of the gene product. It is exported to the EMBL output of the genome. This is not the case for any entries in the Comment field.
Below the description field there is the check box 'Experimental' which should be clicked if any of the results of the annotation are verified in the wet lab. Following are entries concerning the COG (Cluster of orthologous group) functional classification, namely the COG number, COG funccat ID and COG funccat Description. These fields cannot be edited manually, but only pressing the button 'Get' on the right. In the upcoming dialog a COG number can be selected and the entries will be filled.
In the 'Function' field a short description of the function of the gene can be inserted. A confidence for the complete annotation is also to be entered in the respective drop down field. There are different confidence stati that can be chosen here. The strongest confidence is 'High confidence in function and specificity' and the weakest is 'Hypothetical protein'.
The following fields have already been described for the Region Annotation.