GenDBWiki/ToolAndJobConcept
The GenDB Tool and Job Concept
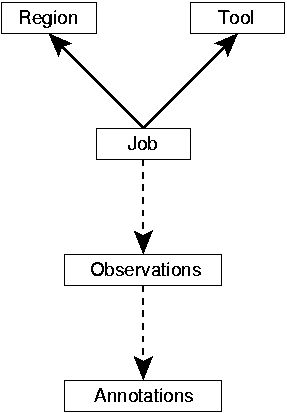
One major improvement of the GenDB system in comparison to the first version, is the modular concept for the integration of bioinformatics tools (e.g. Blast). GenDB allows the incorporation of arbitrary programs for different kinds of bioinformatics analysis. According to the system design, each of these programs is integrated as a Tool (e.g. Tool::Function::Blast), which creates Observations for a specific kind of Region. A Job that can be submitted to the scheduling system thus contains the information about a valid tool and region combination as illustrated below.
For most tools, GenDB also features simple automatic annotators that can be activated. They are started upon completion of a tool run and create automatic annotations employing a simple "best hit" strategy based on the observations created by the tool run.
For an automated large scale computation of various bioinformatics tools, a scalable framework was developed and implemented which allows a batch submission of thousands of Jobs in a very simple manner. Therefore, the following steps have to be performed:
1. The desired Jobs have to be created, e.g. for region or function prediction by using the JobSubmitter Wizard. This can be done quite easily with the submit_job.pl script or via the graphical user interface. For all valid region and tool combinations as defined by the user, the requested Jobs will be created and stored in the Gen``DB project database. Initially, these new Jobs will then have the status PENDING.
2. Before the submit_job.pl script finishes, it calls the submit method of the JobSubmitter Wizard. Thus, all previously created Jobs will be registered as a Job Array in the Scheduler::Codine using the Scheduler::Codine->freeze method. Finally, the array of all Jobs is submitted by calling Scheduler::Codine->thaw. All Jobs should now have the status SUBMITTED and a queue of Jobs should appear in the status report of the Sun GridEngine's qstat output.
3. In the previous step, each Job was submitted to the scheduler by adding the command line for each single Job computation to the list of Jobs. Actually, the script runtool.pl is called for each Job with the corresponding arguments such as runtool.pl -p <projectname> -j <jobid> [-a].
4. When such a command line is executed by one of the compute hosts, the script runtool.pl tries to initialize the Job object for the given id and project name. Since a Job contains the information about a specific region and a single tool that should be computed for that region, this script can now execute the run method that has to be defined for each tool. Such a run method normally starts a bioinformatics tool (e.g. Blast, Pfam, InterPro) for the given region and stores some observations for the results obtained. During this computation the status of the current Job is RUNNING. If the option -a was specified an automatic annotation will be started upon successful computation of the tool. These are only very simple automatic annotations since they are based on the results of a single tool and region combination. Whenever the computation itself or the automatic annotation fails, the status of a Job is set to FAILED, otherwise the status is FINISHED and the computation is complete.
The inclusion of new tools in GenDB is very easy, with the most time-consuming step typically being the implementation of a parser for the result files. For the prediction of regions, such as coding sequences (CDS) or tRNAs, GLIMMER, CRITICA, tRNAscan-SE, and others have been integrated into the system.
Homology searches on DNA or amino acid level in arbitrary sequence databases can be done using the Blast program suite. In addition to using HMMer for motif searches, we also search the BLOCKS and InterPro databases to classify sequence data based on a combination of different kinds of motif search tools. A number of additional tools have been integrated for the characterization of certain features of coding sequences, such as TMHMM for the prediction of alpha-helical transmembrane regions, SignalP for signal peptide prediction, or CoBias for analyzing trends in codon usage.
Since all tools have to be defined separately for each project, a tool configuration wizard was implemented to support this task.
Whereas some tools only return a numeric score and/or an E-value as a result, other tools like Blast or HMMer additionally provide more detailed information, such as an alignment. Although the complete tool results are available to the annotator, only a minimum data subset is stored in form of observations. Based on this subset, the complete tool result record can be recomputed on demand. Storing only a minimal subset of data reduces the storage demands by two orders of magnitude when compared to the traditional "store everything" approach. Our performance measurements have shown this also to be more time efficient than data retrieval from a disk subsystem for any realistic genome project.