GenDBWiki/WebDocumentation/DialogWindows/BlastInterface
Jump to navigation
Jump to search
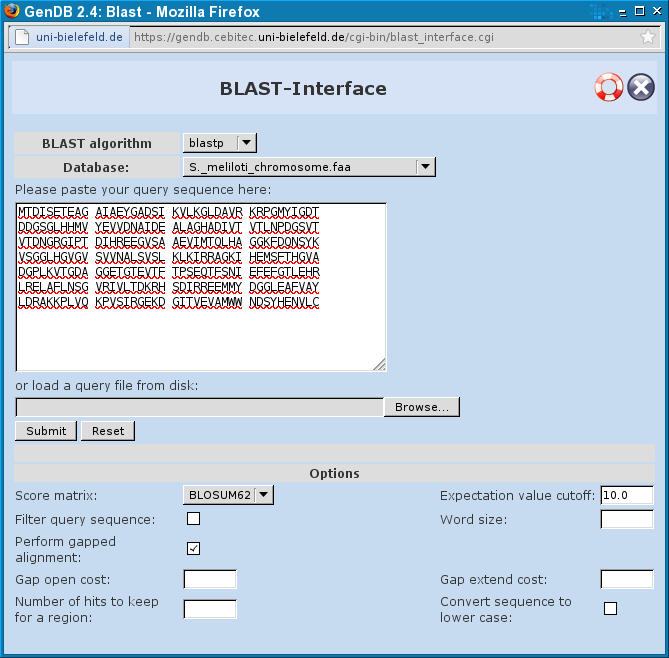
Blast Interface
The Blast Interface can be used to blast DNA or AA sequences vs. global databases (like NR, SWISSPROT...) or vs. private databases like the contigs in the selected project. You can use different Blast-Algorithms here:
- blastp: to blast a protein-sequence vs. a protein-database
- blastn: to blast a DNA-sequence vs. a DNA-database
- blastx: to blast a DNA-sequence vs. a protein-database - the DNA-sequence is translated in all six reading frames
- tblastn: to blast a protein-sequence vs. a DNA-database - the protein database is translated dynamically in all six reading frames
- tblastx: to blast a translated DNA-sequence vs. a translated DNA-database
- psiblast: to blast a protein-sequence vs. a protein-database. psiblast uses position-specific-score-matrices to score matches, it may be more sensitive
You can choose different databases according to the algorithm you selected.
The input-sequence can be copied into the text-field or you can load a query file from your disk.
Other options are explained here.
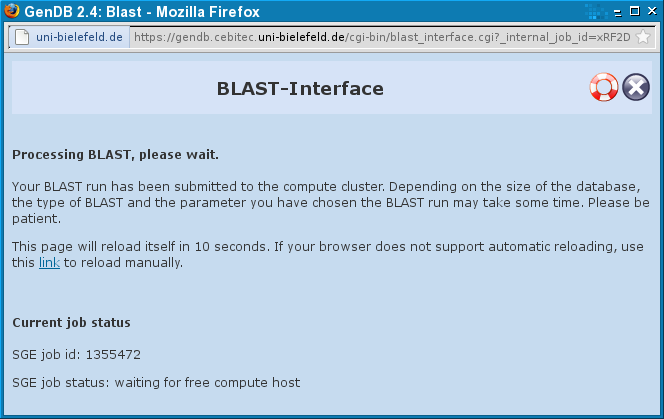
When you made all your choices you can start the BLAST-run by clicking "Submit". You will see the following page as long as the result is being computed: